High-quality images with adequate contrast and details are crucial for many medical imaging applications: e.g., segmentation and computer-aided diagnosis. However, medical images acquired using the same or different sensors usually have a large variation in quality - intensity inhomogeneity, noticeable blur and poor contrast, that are often inherited from the image acquisition process. Cycle-consistent generative adversarial network (CycleGAN) has an advantage of learning knowledge represented with typical images in one domain and transferring it to the other domain without paired images. However, CycleGAN mainly exploits global constraints on appearance and cycle-consistency, which is weak in learning local details. To address the weakness, two novel constraints including an illumination regularization and a structure loss are proposed in our new method, which we refer it to CSI-GAN for medical image enhancement. In our work, low- and high-quality images are treated as those in two different domains and high-quality images can be easily identified by clinicians. The main contributions of this paper are summarized as follows. (1) A novel CSI-GAN is proposed to improve low-quality medical images with better illumination conditions while well-preserving structure details. (2) The proposed method has undergone rigorous quantitative and qualitative evaluation using corneal confocal microscopy and endoscopic images in an unified manner. (3) As a complementary output, we will release the CCM dataset (both poor and good quality image sets) online available to the public in the future.
Proposed Approach
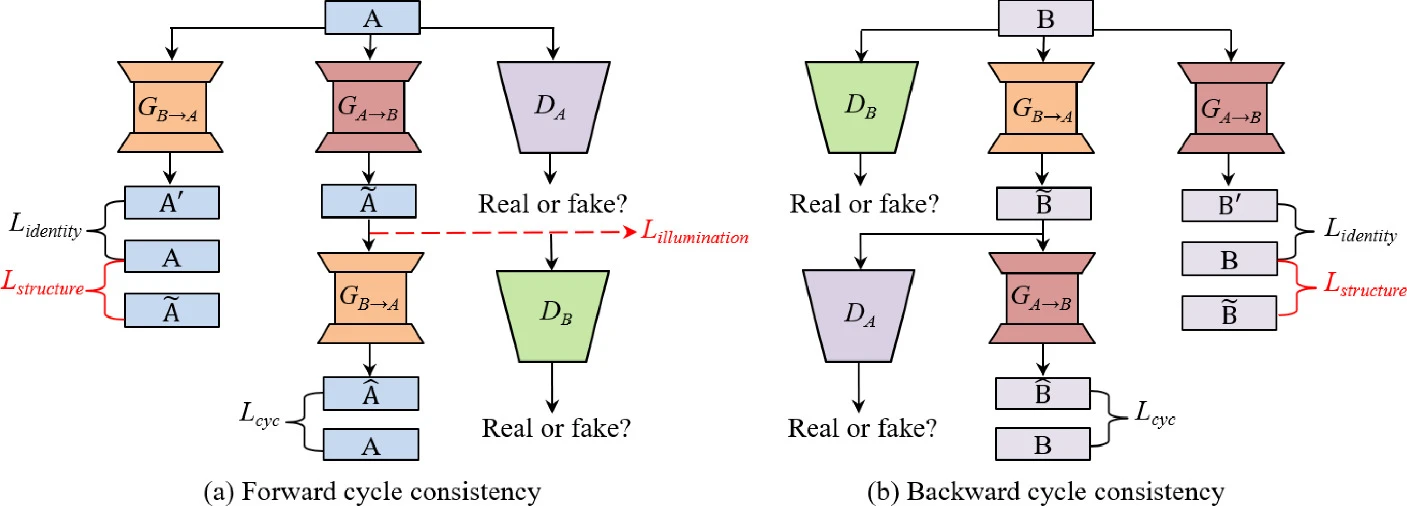
The overview of the CSI-GAN architecture is shown in Figure 1. Different from conditional adversarial network (cGAN), CycleGAN learns a suitable translation function between the source domain A and the target domain B without the paired images in the training. In this paper, we assume that A and B are image domains with low and high quality images, respectively. CycleGAN adopts two generator/discriminator pairs (GA→B/DB, GB→A/DA), where GA→B (GB→A) learns to translate an image from domain A (B) into domain B (A), and DA (DB) is trained to distinguish between real samples from domain A (B) and the translated images from domain B (A). In order to prevent two generators from contradicting each other, the whole framework contains both forward and backward cycle consistency, as shown in Figure 1. Each a∈A is expected to be reconstructed as much as possible in forward cycle, which is represented as a→GA→B(a)→GB→A(GA→B(a))≈a. This holds for backward cycle as well: b→GB→A(b)→GA→B(GB→A(b))≈b. In addition, two generators are regularized as an identity mapping separately when real samples from A (B) are applied to GB→A (GA→B), i.e., GB→A(a)≈a and GA→B(b)≈b.
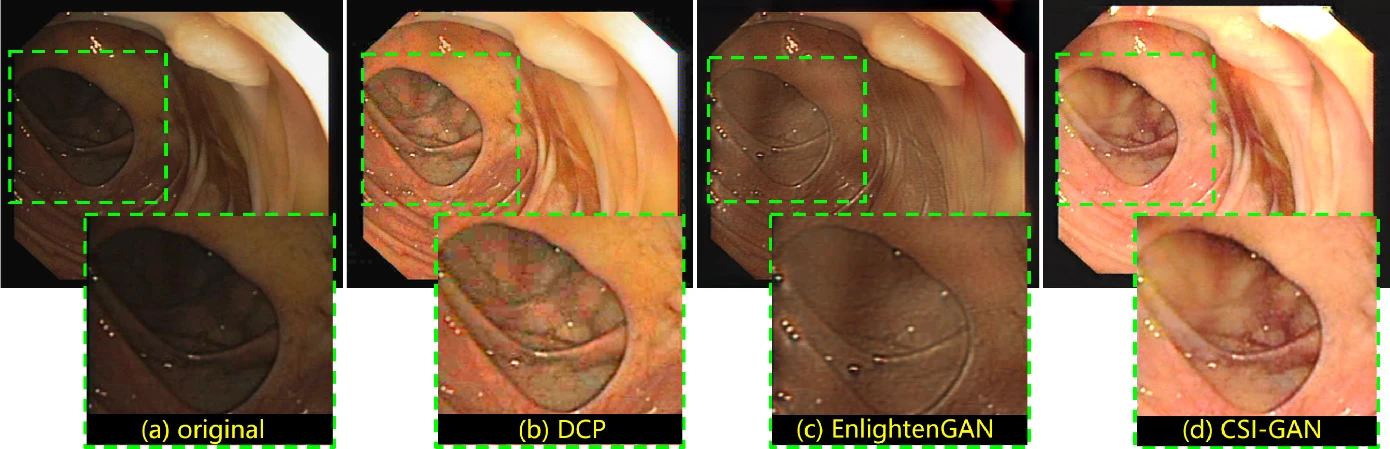
Experimental Results
Evaluation on Endoscopic Images
| Methods | NIQE | BRISQUE | PIQE |
| Original | 4.40±0.67 | 36.70±5.42 | 37.27±11.50 |
| DCP | 4.13±0.64 | 35.45±5.59 | 35.09±6.37 |
| NST | 9.42±1.96 | 30.28±3.64 | 25.48±6.17 |
| MSG-Net | 7.26±0.55 | 56.72±2.29 | 93.43±11.96 |
| EnlightenGAN | 4.38±0.88 | 24.35±4.18 | 33.07±6.47 |
| CycleGAN | 4.38±0.62 | 27.81±7.70 | 29.54±5.28 |
| CSI-GAN (ours) | 3.84±0.64 | 24.52±5.38 | 23.48±6.42 |
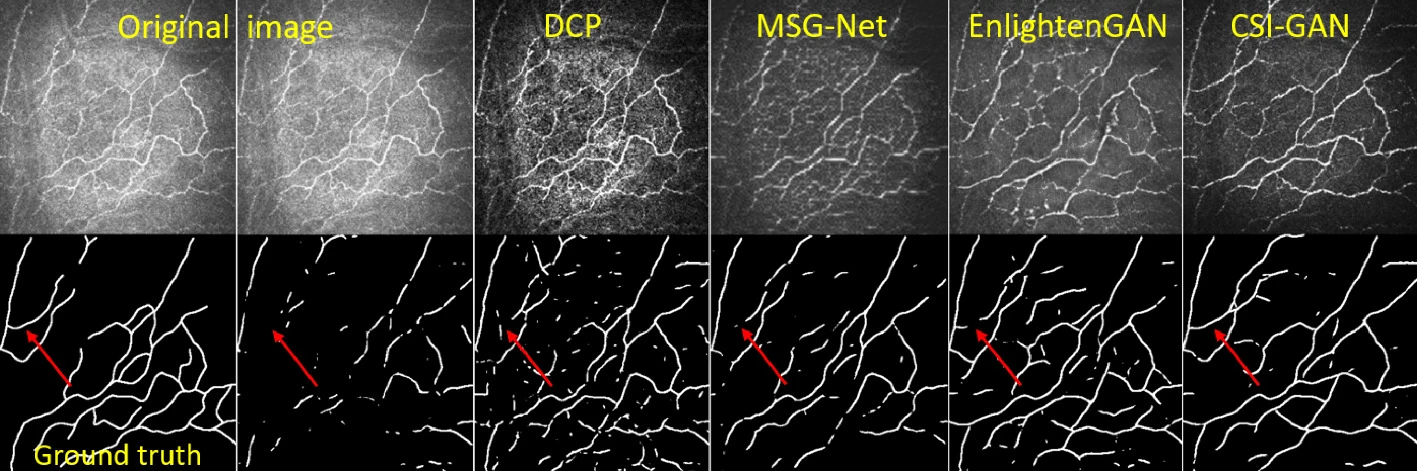
Evaluations on Corneal Confocal Microscopy
| SNR | Segmentation | ||||||
| Methods | r=3 | r=5 | r=7 | ACC | SEN | Kappa | Dice |
| Original | 17.472 | 17.611 | 17.650 | 0.969 | 0.421 | 0.528 | 0.541 |
| CLAHE | 16.560 | 16.733 | 16.793 | 0.970 | 0.488 | 0.570 | 0.584 |
| DCP | 14.587 | 14.879 | 14.986 | 0.964 | 0.708 | 0.615 | 0.633 |
| NST | 16.606 | 16.887 | 17.006 | 0.958 | 0.490 | 0.494 | 0.515 |
| MSG-Net | 19.122 | 19.915 | 20.217 | 0.964 | 0.441 | 0.495 | 0.512 |
| EnlightenGAN | 18.407 | 19.257 | 19.699 | 0.960 | 0.671 | 0.580 | 0.601 |
| CycleGAN | 19.557 | 20.138 | 20.409 | 0.971 | 0.748 | 0.673 | 0.688 |
| CSI-GAN | 20.352 | 21.057 | 21.413 | 0.977 | 0.788 | 0.736 | 0.748 |
Reference
- [1] Cycle Structure and Illumination Constrained GAN for Medical Image Enhancement. MICCAI 2020. paper, arXiv, code & data, ↩


